
Workshop Addresses Wildlife Camera Trap, Ecoacoustic and Drone Data Challenges
Exploreabout Workshop Addresses Wildlife Camera Trap, Ecoacoustic and Drone Data Challenges
As part of our Research Software Agenda for Australia, the ARDC is working with the research community to shape better research software for it to be recognised as a first-class research output. Each month, we talk to a leading research software engineer (RSE), sharing their experience and tips on creating, sustaining and improving software for research.
This month, we spoke with Dr Roozbeh Valavi, a Senior Research Scientist at CSIRO. Roozbeh was recently given the ARDC-sponsored award for New Developers of Open Source Software in Ecology by the Ecological Society of Australia for his work on blockCV, a software package for assessing the accuracy of ecological models.
I have a master’s degree in GIS and remote sensing and a PhD in ecological modelling. During my academic journey, I focused on advancing the methodological aspects of species distribution models (SDMs), in particular assessing and improving their predictive performance, and providing tools and guidelines for their use. I’m particularly motivated to solve environmental problems by applying geospatial analysis, data science and statistical ecological modelling.
Developing new scientific methods often requires writing your own code. Given my commitment to ensuring reproducibility, I found developing research software as one of the most effective ways to share my code and methods with scientific communities, and make a positive contribution to science. I started with the R programming language due to its functionality and popularity in spatial ecological modelling, and then expanded my knowledge to other programming languages such as Python.
I’ve been involved in numerous projects with a strong focus on spatial and ecological modelling (some of which are available on GitHub). While coding played a significant role in these projects, not all of them required the development of software packages. Sometimes I’d use existing software produced by others. Two R packages I’ve developed are blockCV and disdat. Maintained on CRAN, disdat houses species distribution data for various taxa worldwide. It’s primarily for comparing different species distribution modelling methods.
In addition to my work with R packages, I also have experience in Python, developing Shiny web applications and provisioning Amazon Web Services (AWS) servers. I use infrastructure-as-code (IaC) tools like Terraform, Ansible, Docker and GitHub actions to streamline server deployment and management.
The development of the blockCV package began in mid-2017 as part of my PhD research. At that time, I planned to assess and compare the predictive performance of species distribution models (SDMs) using a robust validation approach known as spatial cross-validation. My objective was to determine whether models that perform well with nearby training data maintain their accuracy when applied to distant areas. However, there weren’t many options that cover all the nuances of ecological data, such as sparse or clustered samples, and the use of presence-background data for modelling, so I developed a series of R programming code to do this. In the process, I decided to package the code, making it accessible to the broader ecological modelling community.
My aim was to create a versatile and adaptable tool capable of accommodating various types of ecological data, ensuring that it could serve as a valuable resource for researchers in the field. In essence, blockCV splits data into training and testing folds spatially or environmentally to evaluate spatial ecological models such as SDMs (examples). Its primary goal is to facilitate accurate model evaluation, aiding in the selection of appropriate model structures and improving predictive reliability for enhanced ecological management.
In the process [of developing a series of R programming code to assess and compare the predictive performance of SDMs], I decided to package the code, making it accessible to the broader ecological modelling community.
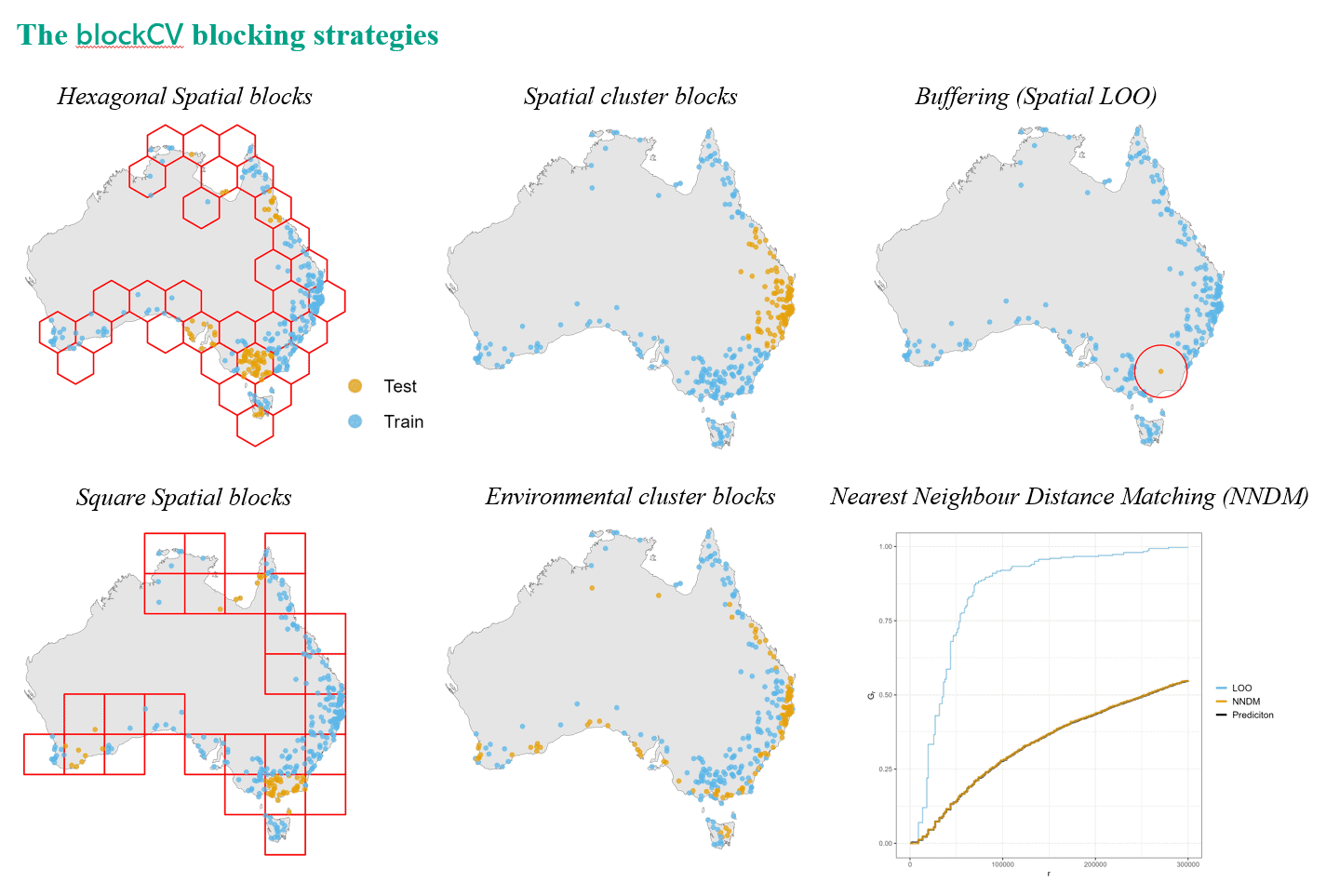
Key features of the blockCV package include the ability to generate train and test folds for k-fold and leave-one-out cross-validations with options for spatial and environmental data separation. It also includes a tool for assessing spatial autocorrelation in response or raster covariates, aiding in the selection of suitable distance bands for data separation. It accommodates diverse SDM scenarios, including presence-absence and presence-background data, rare and common species, and raster-based predictor variables.
The blockCV package has been utilised to evaluate spatial models across diverse fields, including ecology, epidemiology, remote sensing, soil mapping, hydrology and archaeology. Recognising its broader applications beyond ecology, I made significant improvements in the latest major update (v3.0, January 2023). I’ve renamed functions and adjusted arguments to make them more universally understandable, catering to a wider user base while maintaining the functionality of the existing functions. You can learn more on the blockCV GitHub page.
Since its initial release in 2018, the blockCV package has undergone updates and improvements. As of October 2023, it’s been downloaded over 47,000 times from CRAN. The associated paper, published in the journal Methods in Ecology and Evolution, has been cited 350 times, and the package has been adopted by 7 other packages.
I’ve renamed functions and adjusted arguments [for blockCV] to make them more universally understandable, catering to a wider user base while maintaining the functionality of the existing functions.
I was genuinely thrilled and deeply honoured to receive the award. Being recognised for my work in developing research software means a lot to me. Building and maintaining research software comes with significant responsibilities, and when your focus shifts away from the software you’ve created, it becomes even more challenging to keep it up to date and functional. Receiving an award like this serves as a motivating reminder of the importance of open science and research software development. It encourages not only me but also other researchers to actively contribute to the advancement of these valuable initiatives.
Building and maintaining research software comes with significant responsibilities, and when your focus shifts away from the software you’ve created, it becomes even more challenging to keep it up to date and functional.

The R-sig-Geo mailing list is one of my favourites. It’s a fantastic community of R spatial developers who consistently display remarkable responsiveness and willingness to assist others. Another fantastic example is the rOpenSci community, which comes with a lot of resources for R programming.
I recently joined consultations for the ARDC’s Planet Research Data Commons (Planet RDC), an initiative that aims to deliver national datasets and digital resources to facilitate research and informed decision making in environmental and earth sciences. If you have an interest in ecological modelling, I highly recommend exploring EcoCommons, which is supported by ther ARDC. This platform enables access to an extensive database of millions of species records and offers online tools for modelling species distribution under both current conditions and climate change scenarios.
You can connect with Roozbeh via GitHub, LinkedIn and Twitter.
If you’d like to be part of a growing community of RSEs in Australia, become a member of RSE-AUNZ – it’s free!
The ARDC is funded through the National Collaborative Research Infrastructure Strategy (NCRIS) to support national digital research infrastructure for Australian researchers.