
NCRIS Capabilities Seminar: Translational Infrastructure – Working with Researchers and Industry
Go to partner eventabout NCRIS Capabilities Seminar: Translational Infrastructure – Working with Researchers and Industry
As part of our Research Software Agenda for Australia, the ARDC is working with the research community to shape better research software for it to be recognised as a first-class research output. Each month, we talk to a leading research software engineer (RSE), sharing their experience and tips on creating, sustaining and improving software for research.
This month, we spoke with Dr Ryan Wick, a postdoctoral researcher at The Peter Doherty Institute for Infection and Immunity who developed such genomics software as Bandage, Unicycler, Filtlong, Badread, Trycycler and Polypolish. He was awarded the 2023 ARDC-sponsored Australian Bioinformatics and Computational Biology Society (ABACBS) Torsten Seemann Outstanding Bioinformatics Software Developer Award for early- and mid-career researchers.
Ever since I was little, I have been a fan of science. I was a dinosaur kid and a space kid, and I never grew out of it – I still like evolutionary history and space! However, while doing my undergrad in biology, I found that I didn’t enjoy wet lab work. This led to something of an identity crisis: I loved learning science, but I didn’t seem to love doing science, and I was left unsure of what to do with my life.
Many years later, after trying a few different jobs, I took up computer programming as a hobby and really enjoyed it. In hindsight, I should have double-majored biology and computer science. I then learned about the field of bioinformatics, and it seemed a perfect fit for me. I was living in Melbourne (still am), and The University of Melbourne offered a master’s degree in bioinformatics, so I enrolled.
During my master’s and PhD, I was in Kat Holt’s lab, where she supported me to specialise in bacterial genome assembly methods. Over the years, I have encountered a lot of gaps in the genome assembly software ecosystem (either tools that didn’t exist or tools that weren’t good enough), and I try to fill those gaps with tools of my own.
Over the years, I have encountered a lot of gaps in the genome assembly software ecosystem (either tools that didn’t exist or tools that weren’t good enough), and I try to fill those gaps with tools of my own.
In DNA sequencing, where we determine the order in which a DNA molecule’s building blocks come, we use a DNA sequencer to automate the process. DNA sequencers don’t produce whole genomes; they instead make reads or short pieces of the genome, which often contain errors. Putting these sequencing reads back together to get an entire genome sequence is called “assembly”, and the software tools which carry this out are assemblers. Genome assembly is an imperfect process, so the genome sequences used by scientists are often fragmented and contain errors. This has always irked me! It has therefore been my professional mission over the past few years to make assembly of bacterial genomes as accurate as possible – ideally perfect with absolutely no errors at all.
I began using Oxford Nanopore sequencers in 2016, when that platform still had a high error rate. It’s been very fun to witness its development over the last 8 years as its sequencing accuracy has steadily improved. This constant and rapid change has produced many opportunities for software development, and many of my tools involve working with Oxford Nanopore sequencing reads.
Many genome assembly algorithms generate a network-like structure of DNA sequences known as an assembly graph. When I was learning about this topic during my master’s degree, I had assumed there would be a software tool to view assembly graphs. I was surprised to learn that while a few general-purpose graph viewers existed, there was no tool specifically designed for assembly graphs. I spent the next few weeks working on a graphical user interface (GUI) tool that would let me interactively navigate assembly graphs. This grew into Bandage, one of my oldest but most enduring programs. It has been used by researchers in the field of genome assembly for almost 9 years now, including for recent advances in producing a 100% complete human genome.
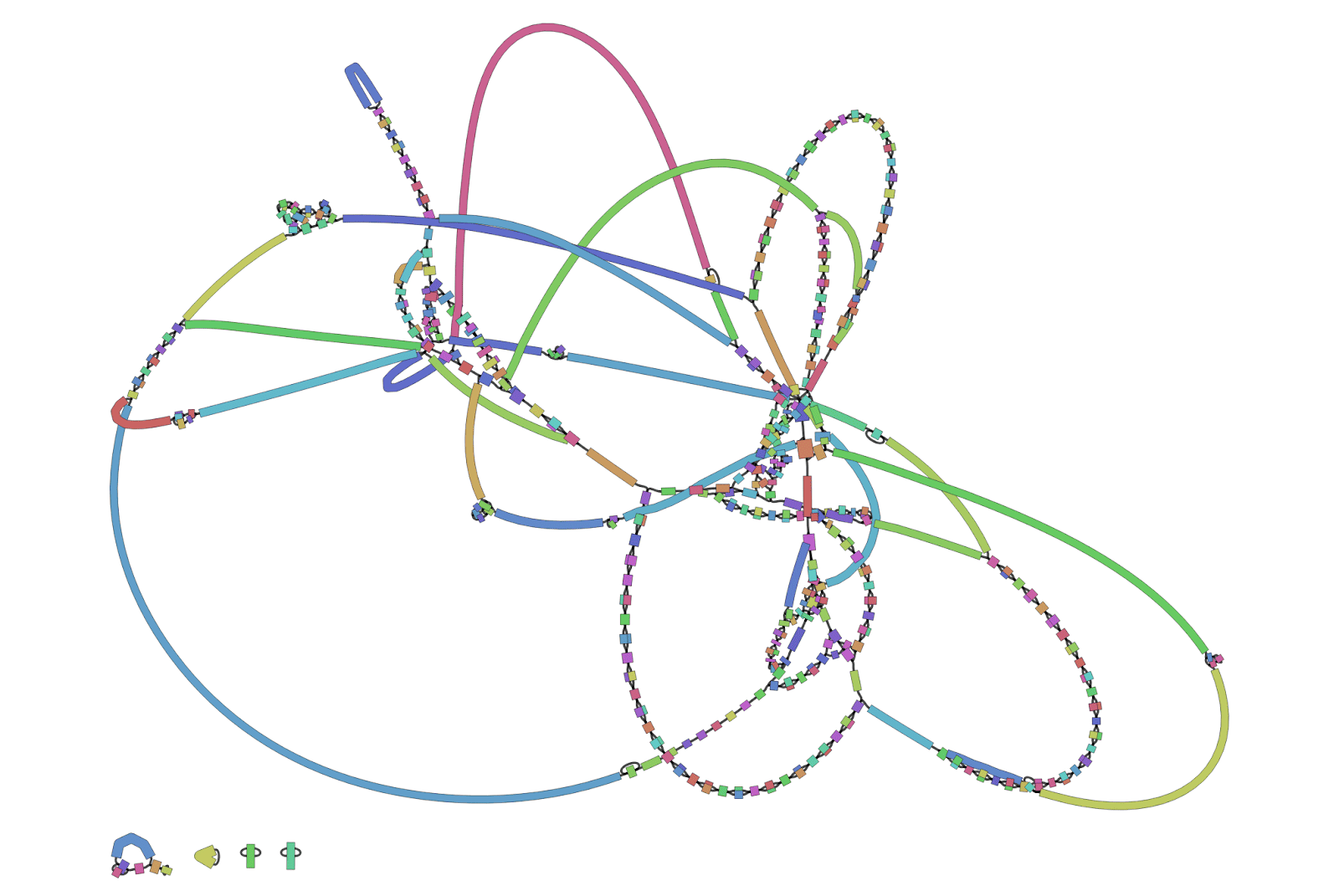
A Bandage visualisation of a bacterial genome assembly graph. Each coloured segment represents a contig, a piece of assembled genomic sequence.
More recently, I’ve been working on genome polishing, a final step in genome assembly where remaining sequence errors are fixed. While there are many software tools for polishing, they often introduce new errors while trying to fix existing errors. It’s like a vacuum cleaner that leaves trails of dirt as it cleans! This led to me developing Polypolish, a genome polishing tool that can fix errors without introducing new ones.
I’m honoured! I recently started working at The Peter Doherty Institute for Infection and Immunity, so Torsten is now a colleague of mine. He seems a little embarrassed that the award is named after him, but he of course deserves it.
Being an RSE can be a lonely endeavour at times. Most of my tools are written by just one person (me), and I know this is also true for many other bioinformatics tools. Software developers in industry often get to code with others (collaborating on a project, pair programming, code review, etc.) and I worry that academic developers like myself are missing out on the many benefits this can bring. I would like to see more cooperative practices in research software development!
Being an RSE can be a lonely endeavour at times. Most of my tools are written by just one person (me), and I know this is also true for many other bioinformatics tools. Software developers in industry often get to code with others (collaborating on a project, pair programming, code review, etc.) and I worry that academic developers like myself are missing out on the many benefits this can bring. I would like to see more cooperative practices in research software development!
I’m a member of the ABACBS. It’s a great way to stay in touch with bioinformaticians that work in different areas. And it’s good to be reminded that there is a lot more to bioinformatics than my niche world of bacterial genome assembly.
You can connect with Ryan via email, GitHub, Mastodon or X/Twitter. He also occasionally posts on his blog.
If you’d like to be part of a growing community of RSEs in Australia, become a member of RSE-AUNZ – it’s free!
The ARDC is proud to sponsor awards for research software and research software engineers in all stages of their careers. The goal of the awards is to strengthen the recognition of research software and those who develop and maintain it as being vital to modern research.
The ARDC continues to sponsor a wide range of research awards for 2024, some of which are now open.
The Astronomical Society of Australia (ASA) has launched the Emerging Leaders in Astronomy Software Development Prize, sponsored by the ARDC.
Friday 16 February 2024
Early-career researchers (ECRs) who have produced or contributed to new astronomy software
Learn more about this award on the ASA website.
The Ecological Society of Australia (ESA) has an ARDC-sponsored award for New Developers of Open Source Software in Ecology.
Sunday 31 March 2024
New software developers from academia, industry or government working on developing open source software for ecology
Learn more about this award on the ESA website.
Sponsored and presented by the ARDC, the Australian Museum Eureka Prize for Excellence in Research Software is awarded for the development, maintenance or extension of software that has enabled significant new scientific research.
7pm (AEST), Friday 12 April 2024
Developers and maintainers of research software
Learn more about this award on the Australian Museum website.
The ARDC is funded through the National Collaborative Research Infrastructure Strategy (NCRIS) to support national digital research infrastructure for Australian researchers.