
NCRIS Facilities Showcase Cutting-Edge Services for Health Researchers and Industry
Exploreabout NCRIS Facilities Showcase Cutting-Edge Services for Health Researchers and Industry
As part of our Research Software Agenda for Australia, the ARDC is working with the research community to shape better research software for it to be recognised as a first-class research output. Each month, we talk to a leading research software engineer (RSE), sharing their experience and tips on creating, sustaining and improving software for research.
This month, we spoke with the predictNMB team’s Rex Parsons. A PhD candidate at the School of Public Health and Social Work of QUT, Rex worked with Robin Blythe, Professor Adrian Barnett, Associate Professor Susanna Cramb and Professor Steven McPhail on predictNMB, a tool for evaluating clinical prediction models based on their net monetary benefit (NMB). Earlier this year, the team was named by the Statistical Society of Australia (SSA) the runner-up for the ARDC-sponsored Venables Award for New Developers of Open Source Software for Data Analytics for developing the tool.
I have a background in biomedical science and biostatistics. I’ve worked in a few different areas of health research and medical device research and development. In academia, I’ve focused on data analysis for circadian rhythms since my honours in 2017 and on clinical prediction models since 2020 when I started my PhD.
My first interest in anything loosely related to being an RSE was out of my honours, when I was first learning R to analyse data from my experiments. I ended up coming up with new methods to fit a nonlinear model to the data and thought that others would be more likely to use it if it were more accessible to those with little to no programming skills. I developed that approach into a pretty rough-looking R package for circadian rhythm analyses, circacompare, and a Shiny app, and it’s since been used quite a bit! Since that time, I’ve been very interested in working with health data and developing software that helps other people do their work better or easier.
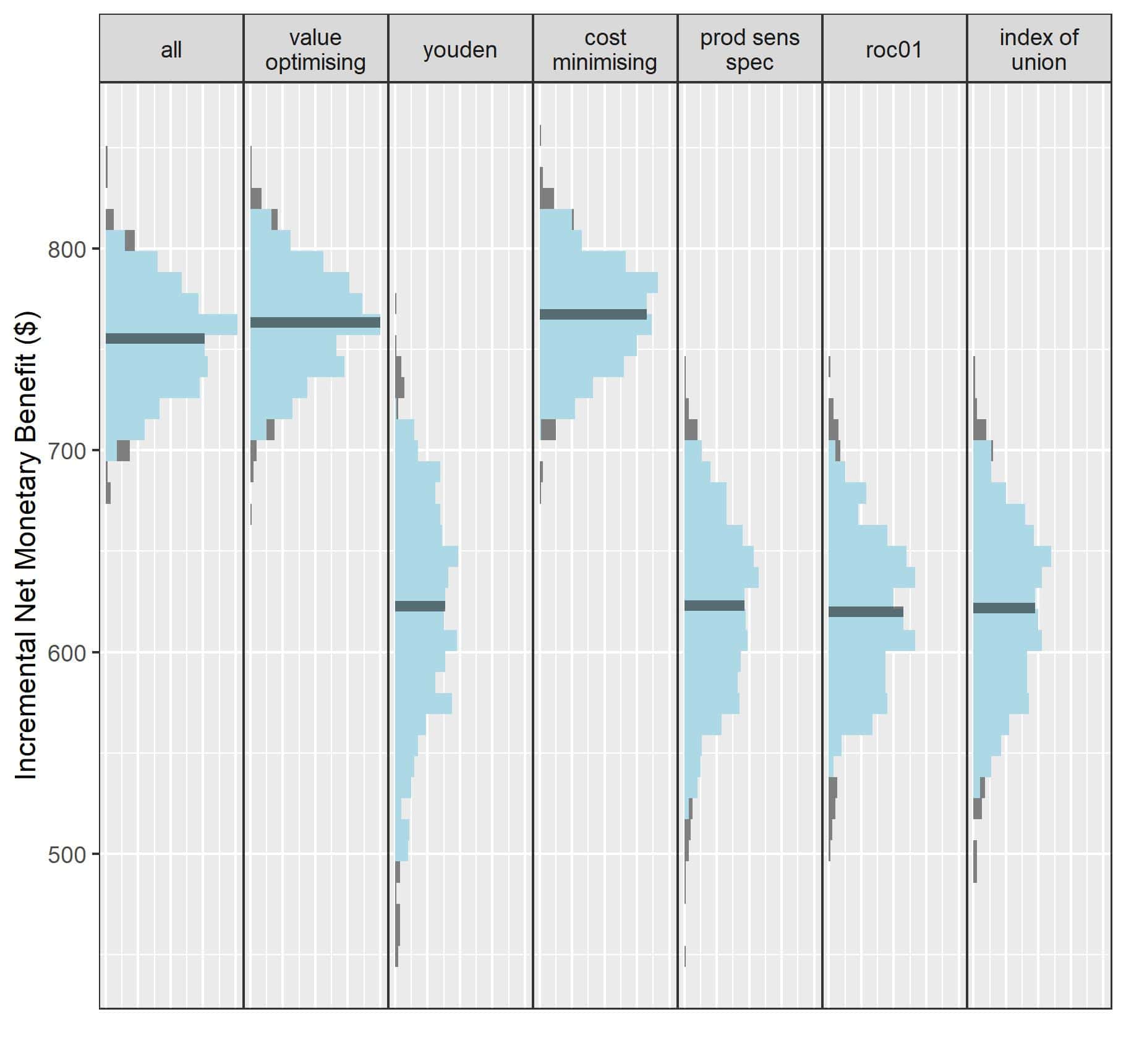
The idea to start this work came out of a study we were doing on cutpoint selection for decision-support systems. We developed a cutpoint selection method which considers the patient and economic outcomes of treatment assigned by a decision-support tool, and we evaluated it with a simulation study. It was during this time that I realised the sensitivity analysis I was performing could be used as an approach to find out when and where hypothetical decision-support systems could be worthwhile. Lots of clinical prediction models are published every year, but it’s rare to see them evaluated or to see whether their predictions will improve decision-making. Wrapping this process up into an R package is what led to predictNMB.
predictNMB can be used to evaluate a real or hypothetical prediction model in health economic terms. This is important because often decision-support tools are only useful if the decision they are guiding can lead to better outcomes. For example, if I have an excellent model that predicts an illness with great accuracy but the intervention for it is ineffective or hugely expensive, the model predictions might be useless! By adjusting the inputs to the functions in predictNMB, you may find that whether models are useful may depend heavily on the treatment available, the costs associated with treatment, or the prevalence of the event being predicted. You might find that you need a model to have impossibly high performance to be better than just treating everyone when the cost of a very effective treatment is almost zero.
The huge gap between the number of prediction models developed versus those implemented and shown to be effective comes at a huge research cost and waste. I hope that people may be inclined to consider what happens after the model makes a prediction and how good their models need to be in the context before developing a model, and predictNMB may help reduce that gap.
I’ve had great feedback on the software from both academics and people working in industry, so I’m hoping they’re finding it useful and using it to make better decisions. Aside from a paper in the Journal of Open Source Software (JOSS), I presented a poster at the International Conference on Health Policy Statistics (ICHPS) in Arizona, US earlier this year and gave a one-hour demo for the R/Medicine conference in August. Robin is also presenting in New Zealand on predictNMB at the end of 2023.
I’m very happy to win such a prestigious award! I felt like the nuts and bolts of predictNMB and the actual simulation were perhaps a bit too basic to merit a statistics software, but I did spend a lot of time trying to make it as usable, documented and tested as possible. I think it was also improved a lot by going through review at rOpenSci and making adjustments to abide by the community’s high standards.
I think it was also improved a lot by going through review at rOpenSci and making adjustments to abide by the community’s high standards.
I did most of the development alongside Robin, and got helpful feedback from all others on the team. I came into this project with next to no experience with health economics, so I’m glad I got a lot of feedback from the team regarding how best to present results from the package functions to the user and the vignettes (i.e. little examples of how the software works).
predictNMB is the first software project that we’ve worked on together as a team, but we are all based at the Australian Centre for Health Services Innovation (AusHSI) at QUT and have worked on other analytical projects. I have worked to develop a Shiny app with Susanna and have worked on reviews for our PhDs with Robin. Individually, I’ve developed circacompare, simMetric (an R package for evaluating simulation studies of statistical methods) and DSSP (an R package to fit Bayesian spatial models with direct sampling).
I’m part of runapp and the rOpenSci Slack workspaces, which are both great and active communities with many RSEs and R experts. I’m also part of the r4ds online learning community, which is much larger and international. I participated in a book club for rpkgs through that community in 2022 and found it really helpful in conceiving predictNMB.
You can connect with the team in the following ways:
If you’d like to be part of a growing community of RSEs in Australia, become a member of RSE-AUNZ – it’s free!
The ARDC is funded through the National Collaborative Research Infrastructure Strategy (NCRIS) to support national digital research infrastructure for Australian researchers.